Unconstrained Adaptable Radial Axes (ARA) mappings using the L-infinity norm
Source:R/ara_unconstrained_linf.R
ara_unconstrained_linf.Rdara_unconstrained_linf() computes unconstrained
Adaptable Radial Axes (ARA) mappings for the L-infinity
norm
Arguments
- X
Numeric data matrix of dimensions N x n, where N is the number of observations, and n is the number of variables.
- V
Numeric matrix defining the axes or "axis vectors". Its dimensions are n x m, where 1<=m<=3 is the dimension of the visualization space. Each row of
Vdefines an axis vector.- weights
Numeric array specifying optional non-negative weights associated with each variable. The function only considers them if they do not share the same value. Default: array of n ones.
- solver
String indicating a package for solving the linear problem(s). It can be "clarabel" (default), "glpkAPI", "Rglpk", or "CVXR".
- use_glpkAPI_simplex
Boolean parameter that indicates whether to use the simplex algorithm (if
TRUE) or an interior point method (ifFALSE), when using the glpkAPI solver. The default isTRUE.- cluster
Optional cluster object related to the parallel package. If supplied, and
n_LP_problemsis N, the method computes the mappings using parallel processing.
Value
A list with the three following entries:
PA numeric N x m matrix containing the mapped points. Each row is the low-dimensional representation of a data observation in X.
statusA vector of length N where the i-th element contains the status of the chosen solver when calculating the mapping of the i-th data observation. The type of the elements depends on the particular chosen solver.
objvalThe numeric objective value associated with the solution to the optimization problem, considering matrix norms, and ignoring weights.
If the chosen solver fails to map one or more data observations (i.e., fails
to solve the related optimization problems), their rows in P will
contain NA (not available) values. In that case, objval will
also be NA.
Details
ara_unconstrained_linf() computes low-dimensional point
representations of high-dimensional numerical data (X) according to
the data visualization method "Adaptable Radial Axes" (M. Rubio-Sánchez,
A. Sanchez, and D. J. Lehmann (2017), doi: 10.1111/cgf.13196),
which describes a collection of convex norm optimization problems aimed at
minimizing estimates of original values in X through dot products of
the mapped points with the axis vectors (rows of V). This particular
function solves the unconstrained optimization problem in Eq. (10), for the
L-infinity vector norm. Specifically, it solves equivalent linear problems as
described in (12). Optional non-negative weights (weights) associated
with each data variable can be supplied to solve the problem in Eq. (15).
References
M. Rubio-Sánchez, A. Sanchez, D. J. Lehmann: Adaptable radial axes plots for improved multivariate data visualization. Computer Graphics Forum 36, 3 (2017), 389–399. doi:10.1111/cgf.13196
Examples
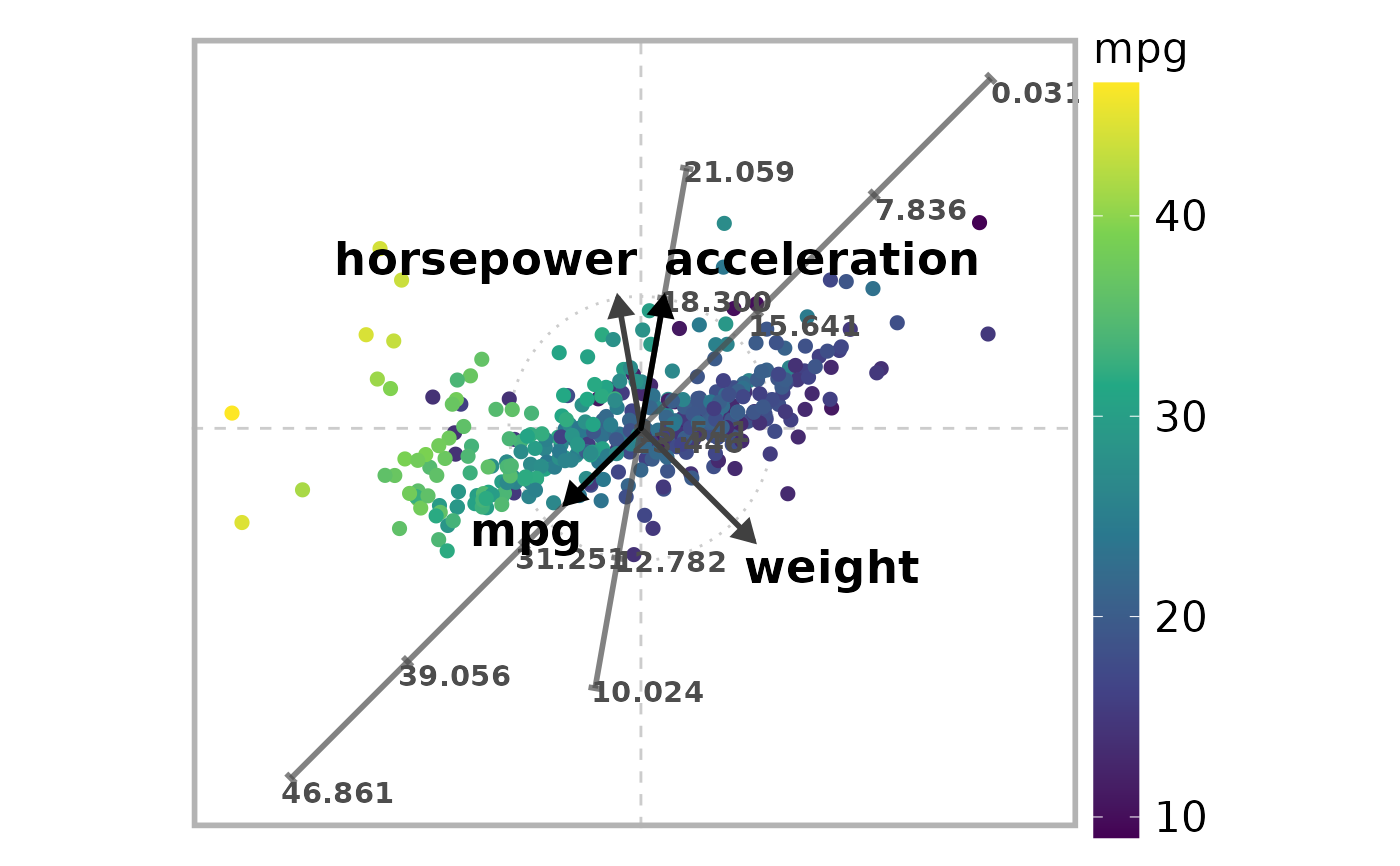
# Define subset of (numerical) variables
# 1:"mpg", 4:"horsepower", 5:"weight", 6:"acceleration"
selected_variables <- c(1, 4, 5, 6)
n <- length(selected_variables)
# Retain only selected variables and rename dataset as X
X <- auto_mpg[, selected_variables] # Select a subset of variables
# Remove rows with missing values from X
N <- nrow(X)
rows_to_delete <- NULL
for (i in 1:N) {
if (sum(is.na(X[i, ])) > 0) {
rows_to_delete <- c(rows_to_delete, -i)
}
}
X <- X[rows_to_delete, ]
# Convert X to matrix
X <- apply(as.matrix.noquote(X), 2, as.numeric)
# Standardize data
Z <- scale(X)
# Define axis vectors (2-dimensional in this example)
r <- c(0.8, 1, 1.2, 1)
theta <- c(225, 100, 315, 80) * 2 * pi / 360
V <- pracma::zeros(n, 2)
for (i in 1:n) {
V[i,1] <- r[i] * cos(theta[i])
V[i,2] <- r[i] * sin(theta[i])
}
# Define weights
weights <- c(1, 0.75, 0.75, 1)
# Set the number of CPU cores/workers
# NCORES <- parallelly::availableCores(omit = 1)
# NCORES <- max(1,parallel::detectCores() - 1)
NCORES <- 2L
# Create a cluster for parallel processing
cl <- parallel::makeCluster(NCORES)
# Compute the mapping
mapping <- ara_unconstrained_linf(
Z,
V,
weights = weights,
solver = "glpkAPI",
use_glpkAPI_simplex = TRUE,
cluster = cl
)
# Stop cluster
parallel::stopCluster(cl)
# Select variables with labeled axis lines on ARA plot
axis_lines <- c(1, 4) # 1:"mpg", 4:"acceleration")
# Select variable used for coloring embedded points
color_variable <- 1 # "mpg"
# Draw the ARA plot
draw_ara_plot_2d_standardized(
Z,
X,
V,
mapping$P,
weights = weights,
axis_lines = axis_lines,
color_variable = color_variable
)